# get OSM data
library(osmar)
library(maptools)
rotewandsteig <- get_osm(way(166274005), full = T)
sp_rotewandsteig <- as_sp(rotewandsteig, what = "lines")
# convert to KML
kmlLine(sp_rotewandsteig@lines[[1]], kmlfile = "rotewandsteig.kml",
lwd = 3, col = "blue", name = "Rotewandsteig")
# view it
shell.exec("rotewandsteig.kml")
26 Sept 2014
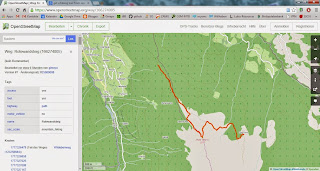
Make a KML-File from an OpenStreetMap Trail
Ever wished to use a trail on OSM on your GPS or smartphone? With this neat little R-Script this can easily be done. You'll just need to search OpenStreetMap for the ID of the trail (way), put this as argument to osmar::get_osm, convert to KML and you're good to go!
4 May 2014
R GIS: Function to Reverse KML Paths
This is a function I wrote up for reversing KML-paths. The paths within a KML can be partially matched by their name-tags
## name: ReverseKmlPath
## use: Reverse KML-pathsby matching their Name tags
## arguments: PATH_TO_DOC, the path to the KML-file
## NAME, the value of the name tag, function uses partial matching!
## 'Trail_xyz' will be matched by 'rail'
## requirements: KML-structure with Placemarks containing a and a tag
## author: Kay Cichini
## date: 01-05-2014
## license: CC-BY-NC-SA
ReverseKmlPath <- function(PATH_TO_DOC, NAMES) {
require(XML)
doc <- xmlInternalTreeParse(PATH_TO_DOC)
if (xmlNamespaceDefinitions(doc)[[1]]$uri == "http://www.opengis.net/kml/2.2") {
namespaces <- c(kml = "http://www.opengis.net/kml/2.2")
flag <- 1
} else {
if (xmlNamespaceDefinitions(doc)[[1]]$uri == "http://earth.google.com/kml/2.0") {
namespaces <- c(kml0 = "http://earth.google.com/kml/2.0")
flag <- 0
} else {
stop ("Stopped!: Check namespace issue..")
}
}
for (NAME in NAMES) {
if (flag) {
query <- paste0("//kml:Placemark[contains(kml:name,'", sprintf("%s", NAME), "'", ")]//kml:coordinates")
} else {
query <- paste0("//kml0:Placemark[contains(kml0:name,'", sprintf("%s", NAME), "'", ")]//kml0:coordinates")
}
coords <- tryCatch(getNodeSet(doc, query, namespaces),
error = function(e) message(paste("\nError: *", NAME, "* was NOT successfully matched\n")))
for (i in length(coords)) {
#grab coordinates from node and reverse order
rev_coord_vector <- rev(unlist(strsplit(gsub("\\t|\\n", "", xmlValue(coords[[i]])), "\\s")))
rev_coord_string <- paste(rev_coord_vector, collapse = " ")
# re-insert reversed line-string:
xmlValue(coords[[i]]) <- rev_coord_string
# message
if (flag) {
query <- paste0("//kml:Placemark[contains(kml:name,'", sprintf("%s", NAME), "'", ")]//kml:name")
} else {
query <- paste0("//kml0:Placemark[contains(kml0:name,'", sprintf("%s", NAME), "'", ")]//kml0:name")
}
match <- xmlValue(getNodeSet(doc, query, namespaces)[[i]])
message(paste0("matched name: ", match, "\n..."))
}
}
# save:
message("Reversed paths saved to:")
saveXML(doc, paste0(dirname(PATH_TO_DOC), "/reversed_", basename(PATH_TO_DOC)),
prefix = newXMLCommentNode("This file was created with the R-package XML::saveXML, see: "))
}
## not run:
tf <- tempfile(fileext = ".kml")
download.file("http://dev.openlayers.org/releases/OpenLayers-2.13.1/examples/kml/lines.kml", tf, mode = "wb")
ReverseKmlPath( PATH_TO_DOC = tf, NAMES = c("Absolute", "Relative") )
shell.exec(tf)
shell.exec(paste0(dirname(tf), "/reversed_", basename(tf)))
3 May 2014
R GIS: Generalizer for KML Paths
I'm posting a recent project's spin-off, which is a custom line-generalizer which I used for huge KML-paths. Anyone with a less clumpsy approach?
## line generalizing function: takes two vectors of with x/ycoords
## and return ids of x/y elements which distance to its next element
## is shorter than the average distance between consecutive vertices
## multiplied by 'fac'
check_dist <- function(x, y, fac) {
dm <- as.matrix(dist(cbind(x, y)))
## supradiagonal holds distance from 1st to 2nd, 2nd to 3rd, etc. element
d <- diag(dm[-1, -ncol(dm)])
mean_dist <- mean(d)
keep <- logical()
## allways keep first..
keep[1] <- T
for (i in 1:(length(x) - 2)) {
keep[i + 1] <- (d[i] > mean_dist * fac)
message(paste0("Distance from item ", i, " to item ", i + 1, " is: ", d[i]))
}
message(paste0("Treshold is: ", mean_dist * fac))
cat("--\n")
## .. and always keep last
keep[length(x)] <- T
return(keep)
}
## Testing function check_dist:
x <- rnorm(5)
y <- rnorm(5)
(keep <- check_dist(x, y, 1.2))
plot(x, y)
lines(x[keep], y[keep], lwd = 4, col = "green")
lines(x, y, lwd = 1, col = "red")
text(x, y + 0.1, labels = c(1:length(x)))
## exclude vertices by generalization rule. coordinate-nodes with low number of vertices,
## segments with less than 'min_for_gen' vertices will not be simplified, in any case coordinates will be
## rounded to 5-th decimal place
generalize_kml_contour_node <- function(node, min_for_gen, fac) {
require(XML)
LineString <- xmlValue(node, trim = T)
LineStrSplit <- strsplit(unlist(strsplit(LineString, "\\s")), ",")
# filter out empty LineStrings which result from strsplit on '\\s'
LineStrSplit <- LineStrSplit[sapply(LineStrSplit, length) > 0]
# all 3 values are required, in case of error see for missing z-values:
x <- round(as.numeric(sapply(LineStrSplit, "[[", 1, simplify = T)), 5)
y <- round(as.numeric(sapply(LineStrSplit, "[[", 2, simplify = T)), 5)
z <- round(as.numeric(sapply(LineStrSplit, "[[", 3, simplify = T)), 5)
# for lines longer than 'min_for_gen' vertices, generalize LineStrings
if (length(x) >= min_for_gen) {
keep <- check_dist(x, y, fac)
x <- x[keep]
y <- y[keep]
z <- z[keep]
xmlValue(node) <- paste(paste(x, y, z, sep = ","), collapse = " ")
# for all other cases, insert rounded values
} else {
xmlValue(node) <- paste(paste(x, y, z, sep = ","), collapse = " ")
}
}
## mind to use the appropiate namespace definition: alternatively use:
## c(kml ='http://opengis.net/kml/2.2')
kml_generalize <- function(kml_file, min_for_gen, fac) {
doc <- xmlInternalTreeParse(kml_file)
nodes <- getNodeSet(doc, "//kml:LineString//kml:coordinates", c(kml = "http://earth.google.com/kml/2.0"))
mapply(generalize_kml_contour_node, nodes, min_for_gen, fac)
saveXML(doc, paste0(dirname(kml_file), "/simpl_", basename(kml_file)))
}
## get KML-files and generalize them
kml_file <- tempfile(fileext = ".kml")
download.file("http://dev.openlayers.org/releases/OpenLayers-2.13.1/examples/kml/lines.kml",
kml_file, mode = "wb")
kml_generalize(kml_file, 5, 0.9)
shell.exec(kml_file)
shell.exec(paste0(dirname(kml_file), "/simpl_", basename(kml_file)))17 Mar 2014
Download all Documents from Google Drive with R
A commentator on my blog recently asked if it is possible to retrieve all direct links to your Google Documents. And indeed it can be very easily done with R, just like so:
# you'll need RGoogleDocs (with RCurl dependency..)
install.packages("RGoogleDocs", repos = "http://www.omegahat.org/R", type="source")
library(RGoogleDocs)
gpasswd = "mysecretpassword"
auth = getGoogleAuth("kay.cichini@gmail.com", gpasswd)
con = getGoogleDocsConnection(auth)
CAINFO = paste(system.file(package="RCurl"), "/CurlSSL/ca-bundle.crt", sep = "")
docs <- getDocs(con, cainfo = CAINFO)
# get file references
hrefs <- lapply(docs, function(x) return(x@access["href"]))
keys <- sub(".*/full/.*%3A(.*)", "\\1", hrefs)
types <- sub(".*/full/(.*)%3A.*", "\\1", hrefs)
# make urls (for url-scheme see: http://techathlon.com/download-shared-files-google-drive/)
# put format parameter for other output formats!
pdf_urls <- paste0("https://docs.google.com/uc?export=download&id=", keys)
doc_urls <- paste0("https://docs.google.com/document/d/", keys, "/export?format=", "txt")
# download documents with your browser
gdoc_ids <- grep("document", types)
lapply(gdoc_ids, function(x) shell.exec(doc_urls[x]))
pdf_ids <- grep("pdf", types, ignore.case = T)
lapply(pdf_ids, function(x) shell.exec(pdf_urls[x]))
3 Mar 2014
Use Case: Make Contour Lines for Google Earth with Spatial R
Here's comes a script I wrote for creating contour lines in KML-format to be used with Google Earth http://github.com/gimoya/theBioBucket-Archives/blob/master/R/contours_for_google_earth.RIf you want to check or just use the datasets I created for the Alps region, you can download it here: http://terrain-overlays.blogspot.co.at/index.html
1 Mar 2014
Use GDAL from R Console to Split Raster into Tiles
When working with raster datasets I often encounter performance issues caused by the large filesizes. I thus wrote up a little R function that invokes gdal_translate which would split the raster into parts which makes subsequent processing more CPU friendly. I didn't use built-in R functions simply because performance is much better when using gdal from the command line..
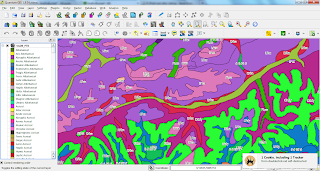
The screenshot to the left shows a raster in QGIS that was split into four parts with the below script.
The screenshot to the left shows a raster in QGIS that was split into four parts with the below script.
## get filesnames (assuming the datasets were downloaded already.
## please see http://thebiobucket.blogspot.co.at/2013/06/use-r-to-bulk-download-digital.html
## on how to download high-resolution DEMs)
setwd("D:/GIS_DataBase/DEM")
files <- dir(pattern = ".hgt")
## function for single file processing mind to replace the PATH to gdalinfo.exe!
## s = division applied to each side of raster, i.e. s = 2 gives 4 tiles, 3 gives 9, etc.
split_raster <- function(file, s = 2) {
filename <- gsub(".hgt", "", file)
gdalinfo_str <- paste0("\"C:/OSGeo4W64/bin/gdalinfo.exe\" ", file)
# pick size of each side
x <- as.numeric(gsub("[^0-9]", "", unlist(strsplit(system(gdalinfo_str, intern = T)[3], ", "))))[1]
y <- as.numeric(gsub("[^0-9]", "", unlist(strsplit(system(gdalinfo_str, intern = T)[3], ", "))))[2]
# t is nr. of iterations per side
t <- s - 1
for (i in 0:t) {
for (j in 0:t) {
# [-srcwin xoff yoff xsize ysize] src_dataset dst_dataset
srcwin_str <- paste("-srcwin ", i * x/s, j * y/s, x/s, y/s)
gdal_str <- paste0("\"C:/OSGeo4W64/bin/gdal_translate.exe\" ", srcwin_str, " ", "\"", file, "\" ", "\"", filename, "_", i, "_", j, ".tif\"")
system(gdal_str)
}
}
}
## process all files and save to same directory
mapply(split_raster, files, 2)
5 Feb 2014
Use Case: Spatial R & Google Earth for Terrain Analyses
I'd like to share code that uses spatial R and Google Earth for terrain analyses. In this example I took SRTM data at 1" resolution from http://www.viewfinderpanoramas.org/dem3.html#alps read it into R did a little processing and finally wrapped it up in a KML-file that I use as ground-overlay in Google Earth. In fact I eventually converted it into a sqlitedb with MAPC2MAPC for usage on a mobile device.
See the code here on github..

See the code here on github..

20 Jan 2014
Get No. of Google Search Hits with R and XML
UPDATE: Thanks to Max Ghenis for updating my R-script which I wrote a while back - the below R-script can now be used again for pulling the number of hits from Google-Search.
GoogleHits <- function(input)
{
require(XML)
require(RCurl)
url <- paste("https://www.google.com/search?q=\"",
input, "\"", sep = "")
CAINFO = paste(system.file(package="RCurl"), "/CurlSSL/ca-bundle.crt", sep = "")
script <- getURL(url, followlocation = TRUE, cainfo = CAINFO)
doc <- htmlParse(script)
res <- xpathSApply(doc, '//*/div[@id="resultStats"]', xmlValue)
cat(paste("\nYour Search URL:\n", url, "\n", sep = ""))
cat("\nNo. of Hits:\n")
return(as.integer(gsub("[^0-9]", "", res)))
}
# Example:
GoogleHits("R%Statistical%Software")
lapply(list_of_search_terms, GoogleHits)
16 Sept 2013
R GIS: Polygon Intersection with gIntersection{rgeos}
A short tutorial on doing intersections in R GIS. gIntersection{rgeos} will pick the polygons of the first submitted polygon contained within the second poylgon - this is done without cutting the polygon's edges which cross the clip source polygon. For the function that I use to download the example data, url_shp_to_spdf() please see HERE.


library(rgeos)
library(dismo)
URLs <- c("http://gis.tirol.gv.at/ogd/umwelt/wasser/wis_gew_pl.zip", # all water bodies in Tyrol
"http://gis.tirol.gv.at/ogd/umwelt/wasser/wis_tseepeicher_pl.zip") # only artificial..
y <- lapply(URLs, url_shp_to_spdf)
z <- unlist(unlist(y))
a <- getData('GADM', country = "AT", level = 2)
b <- a[a$NAME_2=="Innsbruck Land", ] # political district's boundaries
c <- spTransform(b, z[[1]]@proj4string) # (a ring polygon)
z1_c <- gIntersection(z[[1]], c, byid = TRUE)
z2_c <- gIntersection(z[[2]], c, byid = TRUE)
plot(c)
plot(z1_c, lwd = 5, border = "red", add = T)
plot(z2_c, lwd = 5, border = "green", add = T)
plot(z[[1]], border = "blue", add = T) # I plot this on top, so it will be easier to identify
plot(z[[2]], border = "brown", add = T)
Batch Downloading Zipped Shapefiles with R
Here's a function I use to download multiple zipped shapefiles from url and load them to the workspace:
URLs <- c("http://gis.tirol.gv.at/ogd/umwelt/wasser/wis_gew_pl.zip",
"http://gis.tirol.gv.at/ogd/umwelt/wasser/wis_tseepeicher_pl.zip")
url_shp_to_spdf <- function(URL) {
require(rgdal)
wd <- getwd()
td <- tempdir()
setwd(td)
temp <- tempfile(fileext = ".zip")
download.file(URL, temp)
unzip(temp)
shp <- dir(tempdir(), "*.shp$")
lyr <- sub(".shp$", "", shp)
y <- lapply(X = lyr, FUN = function(x) readOGR(dsn=shp, layer=lyr))
names(y) <- lyr
unlink(dir(td))
setwd(wd)
return(y)
}
y <- lapply(URLs, url_shp_to_spdf)
z <- unlist(unlist(y))
# finally use it:
plot(z[[1]])
Follow Up on Spatial Overlays with R - Getting Altitude for a Set of Points
A short follow up on a previous post on spatial overlays with R.


library(sp)
library(dismo)
# some addresses in Austria
pts <- geocode(c("Aldrans, Grubenweg", "Wien, Stephansdom", "Salzburg, Mozartplatz"))
# make pts spatial
coords <- SpatialPoints(pts[, c("longitude", "latitude")])
spdf_pts <- SpatialPointsDataFrame(coords, pts)
# assign CRS/projection (which is WGS 1984)
crs <- CRS(" +proj=longlat +datum=WGS84 +ellps=WGS84 +towgs84=0,0,0")
proj4string(spdf_pts) <- crs
# spatial data to extract (altitude)
alt <- getData('alt', country = "AT")
# convert alt from raster to grid (needed for over::sp)
# and assign CRS (which is the same as spdf_pts, see > alt@crs)
# don't mind warning - the CRS is the same..
spdf_alt <- as(alt, 'SpatialGridDataFrame')
proj4string(spdf_alt) <- crs
# view
plot(alt)
# plot pts on top
plot(spdf_pts, cex = 2, col = 2, add = T)
# check data
str(spdf_pts)
str(spdf_alt)
# get the raster/pixel/grid data (> ?over):
cbind(spdf_pts$interpretedPlace, over(spdf_pts, spdf_alt))
# result:
# spdf_pts$interpretedPlace AUT_msk_alt
# 1 Grubenweg, 6071 Aldrans, Austria 736
# 3 Saint Stephen's Vienna, Stephansplatz 1, 1010 Vienna, Austria 183
# 2 Mozartplatz, 5020 Salzburg, Austria 450
10 Sept 2013
Loading Multiple Shapefiles to the R-Console Simultaneously
A quick tip on how to load multiple shapefiles (point shapefiles, i.e.) to the R console in one go:
library(maptools)
# get all files with the .shp extension from working directory
setwd("D:/GIS_DataBase/GIS_Tirol/Tirol_Verbreitungskarten/Verbreitungs_Daten")
shps <- dir(getwd(), "*.shp")
# the assign function will take the string representing shp and turn it into a variable
# which holds the spatial points data
for (shp in shps) assign(shp, readShapePoints(shp))
plot(get(shp[1])) # i.e.
# ...done
19 Aug 2013
Text Mining with R - Comparing Word Counts in Two Text Documents
Here's what I came up with to compare word counts in two pieces of text. If you got any idea, I'd love to learn about alternatives!
## a function that compares word counts in two texts
wordcount <- function(x, y, stem = F, minlen = 1, marg = F) {
require(tm)
x_clean <- unlist(strsplit(removePunctuation(x), "\\s+"))
y_clean <- unlist(strsplit(removePunctuation(y), "\\s+"))
x_clean <- tolower(x_clean[nchar(x_clean) >= minlen])
y_clean <- tolower(y_clean[nchar(y_clean) >= minlen])
if ( stem == T ) {
x_stem <- stemDocument(x_clean)
y_stem <- stemDocument(y_clean)
x_tab <- table(x_stem)
y_tab <- table(y_stem)
cnam <- sort(unique(c(names(x_tab), names(y_tab))))
z <- matrix(rep(0, 3*(length(cnam)+1)), 3, length(cnam)+1, dimnames=list(c("x", "y", "rowsum"), c(cnam, "colsum")))
z["x", names(x_tab)] <- x_tab
z["y", names(y_tab)] <- y_tab
z["rowsum",] <- colSums(z)
z[,"colsum"] <- rowSums(z)
ifelse(marg == T, return(t(z)), return(t(z[1:dim(z)[1]-1, 1:dim(z)[2]-1])))
} else {
x_tab <- table(x_clean)
y_tab <- table(y_clean)
cnam <- sort(unique(c(names(x_tab), names(y_tab))))
z <- matrix(rep(0, 3*(length(cnam)+1)), 3, length(cnam)+1, dimnames=list(c("x", "y", "rowsum"), c(cnam, "colsum")))
z["x", names(x_tab)] <- x_tab
z["y", names(y_tab)] <- y_tab
z["rowsum",] <- colSums(z)
z[,"colsum"] <- rowSums(z)
ifelse(marg == T, return(t(z)), return(t(z[1:dim(z)[1]-1, 1:dim(z)[2]-1])))
}
}
## example
x = "Hello new, new world, this is one of my nice text documents - I wrote it today"
y = "Good bye old, old world, this is a nicely and well written text document"
wordcount(x, y, stem = T, minlen = 3, marg = T)
Follow-Up:
Thanks a lot for the comments! As I'm not that much into text mining I was trying to reinvent the wheel (in a rather dilettante manner) - missing the capabilities of existing packages. Here's the shortest code that I was able to find doing the same thing (with the potential to get out much more of it, if desired).x = "Hello new, new world, this is one of my nice text documents"
y = "Good bye old, old world, this is a text document"
z = "Good bye old, old world, this is a text document with WORDS for STEMMING - BTW, what is the stem of irregular verbs like write, wrote, written?"
# make a corpus with two or more documents (the cool thing here is that it could be endless (almost) numbers
# of documents to be cross tabulated with the used terms. And the control function enables you
# to do lots of tricks with it before it will be tabulated, see ?termFreq, i.e.)
xyz <- as.list(c(x,y,z))
xyz_corp <- Corpus(VectorSource(xyz))
cntr <- list(removePunctuation = T, stemming = T, wordLengths = c(3, Inf))
as.matrix(TermDocumentMatrix(xyz_corp, control = cntr))
20 Jun 2013
Spatial Overlays with R - Retrieving Polygon Attributes for a Set of Points
A short tutorial for spatial overlays using R-GIS..
library(sp)
library(dismo)
# spatial data (political districts of Austria)
gadm <- getData('GADM', country = "AT", level = 2)
# view
plot(gadm)
# some addresses
pts <- geocode(c("Aldrans, Grubenweg", "Wien, Stephansdom", "Salzburg, Mozartplatz"))
# make pts spatial
coords <- SpatialPoints(pts[, c("longitude", "latitude")])
spdf_pts <- SpatialPointsDataFrame(coords, pts)
# assign CRS/projection (which is WGS 1984)
crs <- CRS(" +proj=longlat +datum=WGS84 +ellps=WGS84 +towgs84=0,0,0")
proj4string(spdf_pts) <- crs
# check data
str(spdf_pts)
str(gadm)
# plot pts on top
plot(spdf_pts, cex = 2, col = 2, add = T)
# do an intersection (points in polygon)
# yielding the polygon's attribute data
over(spdf_pts, gadm)
18 Jun 2013
R GIS: Terrain Analysis for Polygons as Simple as it Gets!

library(rgdal)
library(raster)
alt <- getData('alt', country = "AT")
gadm <- getData('GADM', country = "AT", level = 2)
gadm_sub <- gadm[sample(1:length(gadm), 5), ]
plot(alt)
plot(gadm_sub, add=T)
asp <- terrain(alt, opt = "aspect", unit = "degrees", df = F)
slo <- terrain(alt, opt = "slope", unit = "degrees", df = F)
> extract(slo, gadm_sub, fun = mean, na.rm = T, small = T, df = T)
ID slope
1 1 9.959053
2 2 1.047443
3 3 7.456165
4 4 1.673786
5 5 11.946553
> extract(asp, gadm_sub, fun = mean, na.rm = T, small = T, df = T)
ID aspect
1 1 170.8065
2 2 184.0130
3 3 190.7155
4 4 136.8953
5 5 205.2115
Use R to Bulk-Download Digital Elevation Data with 1" Resolution
Here's a little r-script to convenientely download high quality digital elevation data, i.e. for the Alps, from HERE:
p.s.: Also check raster::getData which pulls SRTM data at 90m resolution for a location / region!
require(XML)
dir.create("D:/GIS_DataBase/DEM/")
setwd("D:/GIS_DataBase/DEM/")
doc <- htmlParse("http://www.viewfinderpanoramas.org/dem3.html#alps")
urls <- paste0("http://www.viewfinderpanoramas.org", xpathSApply(doc,'//*/a[contains(@href,"/dem1/N4")]/@href'))
names <- gsub(".*dem1/(\\w+\\.zip)", "\\1", urls)
for (i in 1:length(urls)) download.file(urls[i], names[i])
# unzip all files in dir and delete them afterwards
sapply(list.files(pattern = "*.zip"), unzip)
unlink(list.files(pattern = "*.zip"))
p.s.: Also check raster::getData which pulls SRTM data at 90m resolution for a location / region!
21 May 2013
R Quick Tip: Shutdown Windows after Script Has Finished
Quite often I have long procedures running and want to do this over night. However, my computer would still be running all night after the script has finished. This is easily circumvented by the following lines that I put at the end of such a script:
# set working dir
# setwd("C:/Users/Kay/Desktop")
# long procedure:
for(i in 1:1e+5) {cat(i); cat("\n..................\n")}
d <- "something"
# save history
savehistory()
# and worspace
save.image()
# then shutdown after 240 s
system("C:/Windows/system32/shutdown.exe -f -s -t 240")
# this would abort the shutdown:
# system("C:/Windows/system32/shutdown.exe -a")
6 May 2013
Creating a QGIS-Style (qml-file) with an R-Script
How to get from a txt-file with short names and labels to a QGIS-Style (qml-file)?
I used the below R-script to create a style for this legend table where I copy-pasted the parts I needed to a txt-file, like for the WRB-FULL (WRB-FULL: Full soil code of the STU from the World Reference Base for Soil Resources). The vector data to which I applied the style is freely available at ESDAC - you just need to submit a form to get access to the data. BTW, thanks to a helping hand on SO.
You can find the QGIS-styler script in theBioBucket-Repository on GitHub.
I used the below R-script to create a style for this legend table where I copy-pasted the parts I needed to a txt-file, like for the WRB-FULL (WRB-FULL: Full soil code of the STU from the World Reference Base for Soil Resources). The vector data to which I applied the style is freely available at ESDAC - you just need to submit a form to get access to the data. BTW, thanks to a helping hand on SO.
You can find the QGIS-styler script in theBioBucket-Repository on GitHub.

21 Apr 2013
Programmatically Download CORINE Land Cover Seamless Vector Data with R
Thanks to a helpful SO-Answer I was able to download all CLC vector data (43 zip-files) programmatically:
require(XML)
path_to_files <- "D:/GIS_DataBase/CorineLC/Seamless"
dir.create(path_to_files)
setwd(path_to_files)
doc <- htmlParse("http://www.eea.europa.eu/data-and-maps/data/clc-2006-vector-data-version-2")
urls <- xpathSApply(doc,'//*/a[contains(@href,".zip/at_download/file")]/@href')
# function to get zip file names
get_zip_name <- function(x) unlist(strsplit(x, "/"))[grep(".zip", unlist(strsplit(x, "/")))]
# function to plug into sapply
dl_urls <- function(x) try(download.file(x, get_zip_name(x), mode = "wb"))
# download all zip-files
sapply(urls, dl_urls)
# function for unzipping
try_unzip <- function(x) try(unzip(x))
# unzip all files in dir and delete them afterwards
sapply(list.files(pattern = "*.zip"), try_unzip)
# unlink(list.files(pattern = "*.zip"))
12 Apr 2013
Download File from Google Drive/Docs Programmatically with R
Following up my lattest posting on how to download files from the cloud with R..
dl_from_GoogleD <- function(output, key, format) {
## Arguments:
## output = output file name
## key = Google document key
## format = output format (pdf, rtf, doc, txt..)
## Note: File must be shareable!
require(RCurl)
bin <- getBinaryURL(paste0("https://docs.google.com/document/d/", key, "/export?format=", format),
ssl.verifypeer = FALSE)
con <- file(output, open = "wb")
writeBin(bin, con)
close(con)
message(noquote(paste(output, "read into", getwd())))
}
# Example:
dl_from_GoogleD(output = "dl_test.pdf",
key = "1DdauvkcVm5XtRBkQIv1na8PeLAwpCBdW8pALCFpRWeM",
format = "pdf")
shell.exec("dl_test.pdf")
library(RCurl)
setwd(tempdir())
destfile = "test_google_docs.csv"
x = getBinaryURL("https://docs.google.com/uc?export=download&id=0B2wAunwURQNsR0I0a0NlQUlJdzA", followlocation = TRUE, ssl.verifypeer = FALSE)
writeBin(x, destfile, useBytes = TRUE)
shell.exec(paste(tempdir(), "/test_google_docs.csv", sep = ""))
Subscribe to:
Comments
(
Atom
)