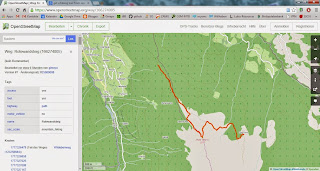
I'm posting a recent project's spin-off, which is a custom line-generalizer which I used for huge KML-paths. Anyone with a less clumpsy approach?
## line generalizing function: takes two vectors of with x/ycoords
## and return ids of x/y elements which distance to its next element
## is shorter than the average distance between consecutive vertices
## multiplied by 'fac'
check_dist <- function(x, y, fac) {
dm <- as.matrix(dist(cbind(x, y)))
## supradiagonal holds distance from 1st to 2nd, 2nd to 3rd, etc. element
d <- diag(dm[-1, -ncol(dm)])
mean_dist <- mean(d)
keep <- logical()
## allways keep first..
keep[1] <- T
for (i in 1:(length(x) - 2)) {
keep[i + 1] <- (d[i] > mean_dist * fac)
message(paste0("Distance from item ", i, " to item ", i + 1, " is: ", d[i]))
}
message(paste0("Treshold is: ", mean_dist * fac))
cat("--\n")
## .. and always keep last
keep[length(x)] <- T
return(keep)
}
## Testing function check_dist:
x <- rnorm(5)
y <- rnorm(5)
(keep <- check_dist(x, y, 1.2))
plot(x, y)
lines(x[keep], y[keep], lwd = 4, col = "green")
lines(x, y, lwd = 1, col = "red")
text(x, y + 0.1, labels = c(1:length(x)))
## exclude vertices by generalization rule. coordinate-nodes with low number of vertices,
## segments with less than 'min_for_gen' vertices will not be simplified, in any case coordinates will be
## rounded to 5-th decimal place
generalize_kml_contour_node <- function(node, min_for_gen, fac) {
require(XML)
LineString <- xmlValue(node, trim = T)
LineStrSplit <- strsplit(unlist(strsplit(LineString, "\\s")), ",")
# filter out empty LineStrings which result from strsplit on '\\s'
LineStrSplit <- LineStrSplit[sapply(LineStrSplit, length) > 0]
# all 3 values are required, in case of error see for missing z-values:
x <- round(as.numeric(sapply(LineStrSplit, "[[", 1, simplify = T)), 5)
y <- round(as.numeric(sapply(LineStrSplit, "[[", 2, simplify = T)), 5)
z <- round(as.numeric(sapply(LineStrSplit, "[[", 3, simplify = T)), 5)
# for lines longer than 'min_for_gen' vertices, generalize LineStrings
if (length(x) >= min_for_gen) {
keep <- check_dist(x, y, fac)
x <- x[keep]
y <- y[keep]
z <- z[keep]
xmlValue(node) <- paste(paste(x, y, z, sep = ","), collapse = " ")
# for all other cases, insert rounded values
} else {
xmlValue(node) <- paste(paste(x, y, z, sep = ","), collapse = " ")
}
}
## mind to use the appropiate namespace definition: alternatively use:
## c(kml ='http://opengis.net/kml/2.2')
kml_generalize <- function(kml_file, min_for_gen, fac) {
doc <- xmlInternalTreeParse(kml_file)
nodes <- getNodeSet(doc, "//kml:LineString//kml:coordinates", c(kml = "http://earth.google.com/kml/2.0"))
mapply(generalize_kml_contour_node, nodes, min_for_gen, fac)
saveXML(doc, paste0(dirname(kml_file), "/simpl_", basename(kml_file)))
}
## get KML-files and generalize them
kml_file <- tempfile(fileext = ".kml")
download.file("http://dev.openlayers.org/releases/OpenLayers-2.13.1/examples/kml/lines.kml",
kml_file, mode = "wb")
kml_generalize(kml_file, 5, 0.9)
shell.exec(kml_file)
shell.exec(paste0(dirname(kml_file), "/simpl_", basename(kml_file)))