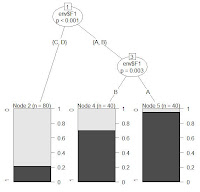
..an illustrative example for testing proportions and presenting the results.
the data: number of indigenous and alien plant species with and without vegetative reproduction (N = 3399, mid-european species, data-courtesy: BiolFlor) . Hypothesis: The proportion of species with vegetative reproduction is different between alien and indigenuos plant species.
result: the prop. of plants with veg. reproduction is sign. lower for alien compared to indigenous plant species. this is simply due to the large number of agricultural weeds and contaminants within alien species - these species almost always reproduce by seeds.
## data:
dat <- data.frame(list(structure(list(flstat = structure(c(2L, 1L, 2L, 1L),
.Label = c("allo", "auto"), class = "factor"),
reprod = structure(c(1L, 1L, 2L, 2L),
.Label = c("non-veg", "veg"), class = "factor"),
X = c(872L, 423L, 1872L, 232L)),
.Names = c("flstat", "reprod", "X"),
class = "data.frame", row.names = c(NA, -4L))))
## proportion of species with vegetative reproduction
p_allo <- dat$X[4] / (dat$X[2] + dat$X[4])
p_auto <- dat$X[3] / (dat$X[1] + dat$X[3])
p_allo
p_auto
## restructure data for glm:
dat1 <- dat[rep(1:4, dat$X), 1:2]
head(dat1)
dat1$inc <- ifelse(dat1$reprod == "non-veg", 0, 1)
## glm:
summary(gmod <- glm(inc ~ flstat, data = dat1, family = binomial))
## intercept = logit(p_allo):
print(est_p_allo <- plogis(gmod$coef[1]))
## intercept + b = logit(p_allo+p_auto):
print(est_p_auto <- plogis(gmod$coef[1] + gmod$coef[2]))
## alternatively test difference in two proportions with prop.test():
ptest_diff <- prop.test(x = c(dat$X[1], dat$X[2]),
n = c(dat$X[1] + dat$X[3], dat$X[2] + dat$X[4]))
## only for one proportion prop.test gives you the confidence
## intervals of p.
## (you could also extract the glm-standard errors and calculate
## the conf.int. for this purpose..):
ptest_auto <- prop.test(x = dat$X[3], n = dat$X[1] + dat$X[3])
ptest_allo <- prop.test(x = dat$X[4], n = dat$X[2] + dat$X[4])
## plot with confidence intervals from prop.test
## (see methods in ?prop.test):
## coordinates for plotting confidence interval bars:
y0_al <- ptest_allo$conf[1]
y1_al <- ptest_allo$conf[2]
y0_au <- ptest_auto$conf[1]
y1_au <- ptest_auto$conf[2]
library(grid)
library(lattice)
## panel function for suppressing tck at top and right side,
## drawing bar with confidence interval,
## plotting glm-estimates (the crosses)
mpanel = function(...) {grid.segments(x0 = c(0.2725, 1 - 0.2725),
x1 = c(0.2725, 1 - 0.2725),
y0 = c(y0_al, y0_au),
y1 = c(y1_al, y1_au))
panel.points(x = c(0.8, 2.2),
y = c(est_p_allo, est_p_auto), pch = 4)
panel.abline(h = c(p_allo, p_auto), lty = 15,
col = "grey70")
panel.text(x = 1.5, y = 0.9, cex = 1.2,
"Species With\nVegetative Reproduction");
panel.xyplot(...)}
xyplot(c(p_allo, p_auto) ~ as.factor(c("Alien", "Indigenous")), type = "b",
ylab = "Prop. +/- CIs\nX = GLM-Estimates",
xlab = "", ylim = c(0, 1),
panel = mpanel, pch = 16,
scales = list(alternating = 1, tck = c(1, 0)))